1.4.3.21: primary-amine oxidase
This is an abbreviated version!
For detailed information about primary-amine oxidase, go to the full flat file.
Reaction
 RCH2NH2 +
RCH2NH2 +
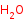 H2O +
H2O +
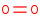 O2=
O2=
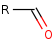 RCHO +
RCHO +
 NH3 +
NH3 +
 H2O2
H2O2
Synonyms
AGAO, AMAO2, AMAO3, amine oxidase 1, amine oxidase, copper containing, AO1, AO2, AOC2, AOC3, BAO, benzylamine oxidase, bovine plasma amine oxidase, bovine serum amine oxidase, BPAO, BSAO, CAO, Copper amine oxidase, copper amine oxidase 1, copper amine oxidase 8, copper-containing amine oxidase, copper-containing AO, copper-containing monoamine oxidase, copper-dependent amine oxidase, copper/quinone containing amine oxidase, Cu/TPQ amine oxidase, CuAO, CuAO1, CuAO2, CuAO3, CuAO8, EC 1.4.3.6, ECAO, ELAO, GPAO, grass pea amine oxidase, Hansenula polymorpha amine oxidase, HPAO, hPAO-1, HPAO-2, lentil seedling amine oxidase, LSAO, MAO-N, More, OVAO, PAO, pea seedling amine oxidase, plasma amine oxidase, PrAO, primary amine oxidase, primary-amine:oxygen oxidoreductase, PSAO, quinone- and copper-containing amine oxidase, quinone-containing copper amine oxidase, quinone-dependent amine oxidase, quinoprotein copper-containing amine oxidase, RAO, sainfoin amine oxidase, semicarbazide-sensitive amine oxidase, semicarbazide-sensitive amine oxidase/vascular adhesion protein-1, SSAO, SSAO/VAP-1, TPQ-containing CuAO, tynA, tyramine oxidase, copper-requiring, VAP-1, vascular adhesion protein 1, vascular adhesion protein-1
ECTree
Sequence
Sequence on EC 1.4.3.21 - primary-amine oxidase
Please wait a moment until all data is loaded. This message will disappear when all data is loaded.
Please wait a moment until the data is sorted. This message will disappear when the data is sorted.
MPO1_TOBAC
790
0
87966
Swiss-Prot
Chloroplast (Reliability: 3)
CUAOZ_ARATH
776
0
86691
Swiss-Prot
Chloroplast (Reliability: 4)
AMO2_ARATH
662
1
75363
Swiss-Prot
Secretory Pathway (Reliability: 1)
AMO2_ARTS1
648
0
72806
Swiss-Prot
-
AMO_ECOLI
Escherichia coli (strain K12)
757
0
84379
Swiss-Prot
-
AOC3_BOVIN
763
1
84500
Swiss-Prot
Secretory Pathway (Reliability: 1)
AOC3_HUMAN
763
1
84622
Swiss-Prot
Secretory Pathway (Reliability: 1)
AOC3_MOUSE
765
1
84534
Swiss-Prot
Secretory Pathway (Reliability: 1)
AOC3_PONAB
763
1
84602
Swiss-Prot
Secretory Pathway (Reliability: 1)
AOC3_RAT
763
1
84981
Swiss-Prot
Secretory Pathway (Reliability: 1)
AOCX_BOVIN
762
0
84757
Swiss-Prot
Secretory Pathway (Reliability: 2)
AOCY_BOVIN
762
0
84883
Swiss-Prot
Secretory Pathway (Reliability: 2)
AOFA_BOVIN
527
1
59758
Swiss-Prot
other Location (Reliability: 2)
AOFA_CANLF
527
1
60088
Swiss-Prot
other Location (Reliability: 2)
AOFA_HORSE
527
1
59580
Swiss-Prot
other Location (Reliability: 2)
AOFA_HUMAN
527
0
59682
Swiss-Prot
other Location (Reliability: 2)
AOFA_MOUSE
526
0
59602
Swiss-Prot
other Location (Reliability: 2)
AOFA_PIG
527
1
59763
Swiss-Prot
other Location (Reliability: 2)
AOFA_PONAB
527
0
59666
Swiss-Prot
other Location (Reliability: 2)
AOFA_RAT
526
0
59508
Swiss-Prot
other Location (Reliability: 2)
AMO1_ARTS1
648
0
72761
Swiss-Prot
-
CAO1_SCHPO
Schizosaccharomyces pombe (strain 972 / ATCC 24843)
712
0
81241
Swiss-Prot
other Location (Reliability: 4)
CAO2_SCHPO
Schizosaccharomyces pombe (strain 972 / ATCC 24843)
794
0
90127
Swiss-Prot
other Location (Reliability: 1)
CAOA2_ARATH
677
0
76674
Swiss-Prot
Secretory Pathway (Reliability: 3)
CAOA3_ARATH
681
0
76711
Swiss-Prot
Secretory Pathway (Reliability: 2)
CAOG1_ARATH
712
1
80138
Swiss-Prot
Secretory Pathway (Reliability: 1)
CAOG2_ARATH
687
1
77533
Swiss-Prot
Secretory Pathway (Reliability: 1)
DAO1_TOBAC
786
0
87421
Swiss-Prot
Chloroplast (Reliability: 4)
PAOX_ARTGO
638
0
70646
Swiss-Prot
-
AMO_KLEAE
755
0
83577
Swiss-Prot
-
AMO_KLEOK
Klebsiella oxytoca (strain ATCC 8724 / DSM 4798 / JCM 20051 / NBRC 3318 / NRRL B-199 / KCTC 1686 / BUCSAV 143 / CCM 1901)
752
0
83624
Swiss-Prot
-
AMO_LENCU
667
0
75558
Swiss-Prot
Secretory Pathway (Reliability: 2)
AMO_PEA
674
1
76358
Swiss-Prot
Secretory Pathway (Reliability: 1)
AMO_PICAN
692
0
77534
Swiss-Prot
Mitochondrion (Reliability: 4)
AO1_ARATH
650
0
73783
Swiss-Prot
Secretory Pathway (Reliability: 2)
AOC2_HUMAN
756
1
83673
Swiss-Prot
Secretory Pathway (Reliability: 1)
AOC2_MOUSE
757
1
83583
Swiss-Prot
Secretory Pathway (Reliability: 1)
AOFA_SHEEP
46
0
5349
Swiss-Prot
other Location (Reliability: 3)
AOFB_BOVIN
520
1
58421
Swiss-Prot
other Location (Reliability: 2)
AOFB_CANLF
520
1
58384
Swiss-Prot
other Location (Reliability: 3)
AOFB_CAVPO
520
0
58410
Swiss-Prot
other Location (Reliability: 3)
AOFB_HUMAN
520
1
58763
Swiss-Prot
other Location (Reliability: 2)
AOFB_MOUSE
520
0
58558
Swiss-Prot
other Location (Reliability: 2)
AOFB_PIG
520
1
58265
Swiss-Prot
other Location (Reliability: 2)
AOFB_PONAB
520
1
58733
Swiss-Prot
other Location (Reliability: 2)
AOFB_RAT
520
1
58459
Swiss-Prot
other Location (Reliability: 2)
AOF_DANRE
522
0
58765
Swiss-Prot
other Location (Reliability: 2)
A0A7H4P4K5_9ENTR
64
0
7784
TrEMBL
-
A0A2X1BAM5_9BACI
328
1
36582
TrEMBL
-
A0A1V5XFX5_9FIRM
295
1
33146
TrEMBL
-
A0A2X3IYD8_KLEPN
125
0
13568
TrEMBL
-
A0A1S1V9S1_9FIRM
429
0
48892
TrEMBL
-
A0A377W288_KLEPN
87
0
9451
TrEMBL
-
A0A377UYW0_KLEPN
238
0
25674
TrEMBL
-
A0A8B8MA99_ABRPR
1389
0
156246
TrEMBL
Secretory Pathway (Reliability: 1)
A0A484Z4Z8_9ENTR
201
0
21916
TrEMBL
-
A0A151AS61_9THEO
162
1
18277
TrEMBL
-
A0A1V4VJA7_9FIRM
221
0
24223
TrEMBL
-
A0A377ZSF1_KLEPO
47
0
5478
TrEMBL
-
A0A7H4SFS6_KLEVA
91
0
9981
TrEMBL
-
A0A1V4W2V1_9FIRM
416
1
44894
TrEMBL
-
A0A7H4LTP0_9ENTR
122
0
13478
TrEMBL
-
A0A099TYQ3_9HELI
466
0
53369
TrEMBL
-
A0A2X3I8G8_KLEPN
82
0
8991
TrEMBL
-
A0A377LFJ2_ECOLX
160
0
17659
TrEMBL
-
A0A377LFU2_ECOLX
63
0
6874
TrEMBL
-
A0A377M3J0_ENTCL
134
0
14943
TrEMBL
-
A0A5D2W5Y7_GOSMU
165
0
18554
TrEMBL
other Location (Reliability: 2)
A0A377UZY1_KLEPN
73
0
8334
TrEMBL
-
A0A5C7IRT7_9ROSI
239
0
27340
TrEMBL
other Location (Reliability: 5)
G8Q1B3_PSEO1
Pseudomonas ogarae (strain DSM 112162 / CECT 30235 / F113)
560
0
62331
TrEMBL
-
A0A377J682_9HELI
61
0
6467
TrEMBL
-
A0A377J510_9HELI
209
0
22990
TrEMBL
-
A0A376YDM2_ECOLX
119
0
13039
TrEMBL
-
A0A853W4J7_MOOTH
162
1
18029
TrEMBL
-
A0A377JSX5_9HELI
537
0
60510
TrEMBL
-
A0A378F4B3_KLEPN
135
0
14384
TrEMBL
-
A0A6P6TZP1_COFAR
616
0
67992
TrEMBL
Mitochondrion (Reliability: 5)
A0A1U8KZW3_GOSHI
175
0
19824
TrEMBL
other Location (Reliability: 2)
A0A376P4Y1_ECOLX
102
0
11220
TrEMBL
-
A0A377TS74_KLEPN
135
0
14507
TrEMBL
-
A0A377ZRW2_KLEPN
184
0
20036
TrEMBL
-
A0A1L1PNJ4_HYDIT
553
0
60347
TrEMBL
-
A0A2X3L4P6_9ENTR
99
0
11002
TrEMBL
-
A0A2X3J3Y4_9ENTR
65
0
7312
TrEMBL
-
A0A151AXG3_9THEO
307
1
32894
TrEMBL
-
A0A377HBC3_ECOLX
52
0
5535
TrEMBL
-
A0A1U8I9B6_GOSHI
175
0
19796
TrEMBL
other Location (Reliability: 2)
A0A2X3CA53_KLEPN
125
0
13748
TrEMBL
-
A0A486P1J3_KLEPN
84
0
9039
TrEMBL
-
G8Q2L0_PSEO1
Pseudomonas ogarae (strain DSM 112162 / CECT 30235 / F113)
589
0
62809
TrEMBL
-
A0A377LHN7_ECOLX
84
0
9225
TrEMBL
-
A0A485D6F8_RAOPL
91
0
10258
TrEMBL
-
A0A377J4K3_9HELI
95
0
10160
TrEMBL
-
A0A7H4MZU2_9ENTR
198
0
21445
TrEMBL
-
A0A377JM22_9HELI
562
0
63443
TrEMBL
-
A0A377TUT3_KLEPN
95
0
10592
TrEMBL
-
A0A4U9HW78_9ENTR
109
0
12223
TrEMBL
-
A0A4V6JJI3_9ENTR
96
0
10547
TrEMBL
-
A0A485D7G8_RAOPL
78
0
9017
TrEMBL
-
A0A485D6R6_RAOPL
70
0
7769
TrEMBL
-
A0A1J5JPG9_MOOTH
298
0
31571
TrEMBL
-
A0A4V6J144_RAOTE
71
0
7615
TrEMBL
-
A0A7H4MZU3_9ENTR
83
0
9058
TrEMBL
-
A0A4U9I1J2_9ENTR
60
0
6464
TrEMBL
-
A0A2J8A092_9CHLO
337
0
34152
TrEMBL
Chloroplast (Reliability: 5)
A0A378EIB3_KLEPR
47
0
5164
TrEMBL
-
A0A0B0IB45_9BACL
532
0
57528
TrEMBL
-
A0A5D2W6D0_GOSMU
175
0
19730
TrEMBL
other Location (Reliability: 3)
A0A378EGV0_KLEPR
54
0
5538
TrEMBL
-
A0A5E6MB27_9BACT
62
0
6809
TrEMBL
-
A0A7H4MZU4_9ENTR
60
0
6609
TrEMBL
-
A0A1E5V3W8_9POAL
440
0
49208
TrEMBL
other Location (Reliability: 4)
A0A448TRB4_9HELI
415
0
47832
TrEMBL
-
M5BLL8_THACB
Thanatephorus cucumeris (strain AG1-IB / isolate 7/3/14)
478
1
52886
TrEMBL
Secretory Pathway (Reliability: 1)
A0A377XRK7_KLEPN
41
0
4250
TrEMBL
-
A0A422MPE6_9TRYP
565
0
59453
TrEMBL
Mitochondrion (Reliability: 3)
A0A2T0ATN8_9THEO
307
1
32963
TrEMBL
-
A0A5B7BX63_DAVIN
618
0
68577
TrEMBL
other Location (Reliability: 2)
A0A377J472_9HELI
611
0
68504
TrEMBL
-
A0A4U9CU11_RAOTE
119
0
12873
TrEMBL
-
A0A2X3B7W1_9HELI
504
0
57314
TrEMBL
-
A0A7H4P4K7_9ENTR
38
0
4209
TrEMBL
-
A0A2X1Q9Q8_KLEPN
51
0
5537
TrEMBL
-
A0A485D604_RAOPL
66
0
7630
TrEMBL
-
A0A377J2S7_9HELI
450
0
49114
TrEMBL
-
A0A0H2LSL1_VARPD
42
0
4680
TrEMBL
-
M5C6B8_THACB
Thanatephorus cucumeris (strain AG1-IB / isolate 7/3/14)
478
1
52895
TrEMBL
Secretory Pathway (Reliability: 1)
I7GYM6_9HELI
614
0
68726
TrEMBL
-
A0A1R3KR96_9ROSI
2841
2
316878
TrEMBL
other Location (Reliability: 2)
A0A377J572_9HELI
1848
0
208279
TrEMBL
-
A0A376ZYN1_ECOLX
118
0
12837
TrEMBL
-
A0A2X1QCM1_KLEPN
138
0
15077
TrEMBL
-
A0A2X1SHB3_KLEPN
135
0
14982
TrEMBL
-
A0A4U9HX25_9ENTR
49
0
5281
TrEMBL
-
A0A5D2M7Y1_GOSTO
175
0
19777
TrEMBL
other Location (Reliability: 2)
A0A377E471_ECOLX
90
0
10092
TrEMBL
-
A0A376MWP5_ECOLX
87
0
9563
TrEMBL
-
A0A5D2DP33_GOSDA
175
0
19796
TrEMBL
other Location (Reliability: 2)
A0A4U9HWA0_9ENTR
168
0
18592
TrEMBL
-
A0A485BGV2_RAOTE
128
0
14036
TrEMBL
-
A0A364NA43_9PLEO
1045
0
117173
TrEMBL
other Location (Reliability: 3)
A0A377V4E1_KLEPN
95
0
10113
TrEMBL
-
A0A4U9HWK8_9ENTR
64
0
7480
TrEMBL
-
A0A2X3J202_9ENTR
66
0
7777
TrEMBL
-
A0A2X1PW89_ECOLX
118
0
12867
TrEMBL
-
A0A853W2E6_MOOTH
298
0
31629
TrEMBL
-
A0A1U8GEV6_CAPAN
1698
1
191824
TrEMBL
Secretory Pathway (Reliability: 1)
A0A1S1VAJ3_9FIRM
380
0
43039
TrEMBL
-
A1R2C3_PAEAT
Paenarthrobacter aurescens (strain TC1)
656
0
72378
TrEMBL
-
A1RDD3_PAEAT
Paenarthrobacter aurescens (strain TC1)
654
0
72450
TrEMBL
other Location (Reliability: 5)
I6NC69_HUPSR
681
0
76854
TrEMBL
-
Q42432_PEA
40
0
4573
TrEMBL
-
Q6A174_LATSA
649
0
73643
TrEMBL
-
Q9SW90_EUPCH
677
0
76510
TrEMBL
-
html completed




 results (
results ( results (
results ( top
top





