1.3.1.3: DELTA4-3-oxosteroid 5beta-reductase
This is an abbreviated version!
For detailed information about DELTA4-3-oxosteroid 5beta-reductase, go to the full flat file.
Reaction
 17,21-dihydroxy-5beta-pregnane-3,11,20-trione +
17,21-dihydroxy-5beta-pregnane-3,11,20-trione +
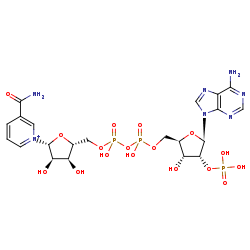 NADP+=
NADP+=
 cortisone +
cortisone +
 NADPH +
NADPH +
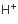 H+
H+
Synonyms
3-oxo-DELTA4-steroid 5beta-reductase, 4,5beta-dihydrocortisone:NADP+ DELTA4-oxidoreductase, 5-beta-reductase, 5beta-POR, 5beta-red, AKR1D1, AKR1D4, aldo-keto reductase, aldo-keto reductase 1D1, aldo�keto reductase 1D1, androstenedione 5beta-reductase, CARUB_v10005016mg, cortisone 5beta-reductase, cortisone beta-reductase, DELTA 4-5beta-reductase, DELTA4-3-ketosteroid 5beta-reductase, DELTA4-3-oxosteroid 5beta-reductase, DELTA4-hydrogenase, EC 1.3.1.23, EC 1.3.1.3, h5beta-red, h5beta-reductase, More, P5betaR, progesterone 5beta-reductase, reductase, cholestenone 5beta.-, reductase, cortisone DELTA4-5beta-, SRD5beta, St5betaR1, steroid 5beta-reductase, steroid 5beta.-reductase, testosterone 5-beta-reductase, testosterone 5beta-reductase
ECTree
Sequence
Sequence on EC 1.3.1.3 - DELTA4-3-oxosteroid 5beta-reductase
Please wait a moment until all data is loaded. This message will disappear when all data is loaded.
Please wait a moment until the data is sorted. This message will disappear when the data is sorted.
5BPOR_DIGLA
389
0
44071
Swiss-Prot
other Location (Reliability: 3)
VEP1_ARATH
388
0
44230
Swiss-Prot
other Location (Reliability: 3)
AK1D1_HUMAN
326
0
37377
Swiss-Prot
other Location (Reliability: 2)
AK1D1_MOUSE
325
0
37290
Swiss-Prot
other Location (Reliability: 2)
AK1D1_RABIT
326
0
37491
Swiss-Prot
other Location (Reliability: 1)
AK1D1_RAT
326
0
37378
Swiss-Prot
other Location (Reliability: 2)
A0A084GGE5_PSEDA
329
0
37042
TrEMBL
other Location (Reliability: 2)
D6P3J0_TOBAC
388
0
43709
TrEMBL
other Location (Reliability: 2)
A0A7D3QMZ6_NEPCA
389
0
44135
TrEMBL
other Location (Reliability: 3)
H6UF83_LUNAN
389
0
43978
TrEMBL
other Location (Reliability: 2)
E7CL26_DIGLA
396
1
44612
TrEMBL
other Location (Reliability: 2)
W8YNV9_9BACL
356
1
39917
TrEMBL
-
A0A075M2G6_9LAMI
389
0
44126
TrEMBL
other Location (Reliability: 3)
B9EMZ1_SALSA
326
0
37403
TrEMBL
other Location (Reliability: 2)
G9HWD8_9LAMI
389
0
44082
TrEMBL
other Location (Reliability: 3)
H2BJQ0_SOLTU
387
0
43650
TrEMBL
other Location (Reliability: 2)
Q5BKI0_XENTR
326
0
37258
TrEMBL
other Location (Reliability: 1)
A0A812EUD9_SEPPH
354
0
38326
TrEMBL
other Location (Reliability: 4)
G9HWD9_9LAMI
389
0
44125
TrEMBL
other Location (Reliability: 3)
A0A061IR69_CRIGR
279
0
31870
TrEMBL
other Location (Reliability: 2)
E3TEK6_ICTPU
326
0
37288
TrEMBL
other Location (Reliability: 1)
G9HWE1_9LAMI
389
0
44112
TrEMBL
other Location (Reliability: 3)
A0A061IQE3_CRIGR
285
0
32684
TrEMBL
other Location (Reliability: 2)
D2JSQ9_DIGPU
394
0
44320
TrEMBL
other Location (Reliability: 2)
H2BJQ1_SOLLC
387
0
43580
TrEMBL
other Location (Reliability: 2)
D9IV94_9ROSI
387
0
43799
TrEMBL
other Location (Reliability: 3)
A0A7D3V2U5_HYSOF
388
0
43894
TrEMBL
other Location (Reliability: 2)
D9ILU8_9SOLA
387
0
43640
TrEMBL
other Location (Reliability: 2)
A0A7D3QN93_NEPCA
388
0
43916
TrEMBL
other Location (Reliability: 3)
A0A075M737_DIGLA
389
0
44159
TrEMBL
other Location (Reliability: 3)
H6UF82_9SOLA
388
0
43487
TrEMBL
other Location (Reliability: 2)
A0A061ILS4_CRIGR
268
0
30621
TrEMBL
other Location (Reliability: 2)
A0A8A9A1F8_BUFGR
326
0
37259
TrEMBL
other Location (Reliability: 1)
G9HWE0_9LAMI
389
0
44127
TrEMBL
other Location (Reliability: 3)
A0A7D3ULA9_NEPRA
389
0
44133
TrEMBL
other Location (Reliability: 3)
Q078S6_9LAMI
389
0
44081
TrEMBL
-
R0F4W3_9BRAS
388
0
44164
TrEMBL
-
html completed



 results (
results ( results (
results ( top
top









