1.13.11.51: 9-cis-epoxycarotenoid dioxygenase
This is an abbreviated version!
For detailed information about 9-cis-epoxycarotenoid dioxygenase, go to the full flat file.
Word Map on EC 1.13.11.51 
Reaction
 9'-cis-neoxanthin +
9'-cis-neoxanthin +
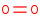 O2=
O2=
 2-cis,4-trans-xanthoxin +
2-cis,4-trans-xanthoxin +
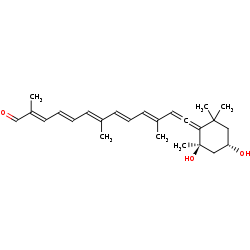 (3S,5R,6R)-5,6-dihydroxy-6,7-didehydro-5,6-dihydro-12'-apo-beta-caroten-12'-al
(3S,5R,6R)-5,6-dihydroxy-6,7-didehydro-5,6-dihydro-12'-apo-beta-caroten-12'-al
Synonyms
9,10[9',10']carotenoid cleavage dioxygenase, 9-cis epoxycarotenoid dioxygenase, 9-cis-epoxcarotenoid dioxygenase, 9-cis-epoxicarotenoid dioxygenase, 9-cis-epoxycarotenoid dioxygenase, 9-cis-epoxycarotenoid dioxygenase 3, 9-cis-epoxycarotenoid dioxygenase 4, 9-cis-epoxycarotenoid dioxygenase 6, 9-cis-epoxycarotenoid dioxygenase1, 9-cis-epoxycarotenoid dioxygenase4, AtCCD1, AtNCED3, B-diox-II, carotenoid cleavage dioxygenase, carotenoid cleavage dioxygenase 1, CCD, CCD1, CCD7, CCD8, CitNCED2, CitNCED3, CMNCED1, CsNCED1, CsNCED2, DHN2, DKNCED1, LeNCED1, LeNCED2, LsNCED1, LsNCED2, LsNCED3, LsNCED4, NCED, NCED 3, NCED1, NCED2, NCED2A, NCED2B, NCED2D, NCED3, NCED3a, NCED4, NCED5, NCED6, NCED9, nine-cis-epoxy dioxygenase, nine-cis-epoxycarotenoid dioxygenase, nine-cis-epoxycarotenoid dioxygenase 3, NosCCD, Nostoc carotenoid cleavage dioxygenase, NSC1, NSC2, NSC3, PpNCED1, PpNCED2, putative carotenoid cleavage dioxygenase, PvNCED1, SgNCED1, STO1, VP14, VP14 epoxy-carotenoid dioxygenase, VP14 protein, VuNCED1 protein, VvNCED1
ECTree
Sequence
Sequence on EC 1.13.11.51 - 9-cis-epoxycarotenoid dioxygenase
Please wait a moment until all data is loaded. This message will disappear when all data is loaded.
Please wait a moment until the data is sorted. This message will disappear when the data is sorted.
NCED1_MAIZE
604
0
65495
Swiss-Prot
Chloroplast (Reliability: 3)
NCED1_ORYSJ
638
0
68620
Swiss-Prot
Chloroplast (Reliability: 3)
NCED1_PHAVU
615
0
68075
Swiss-Prot
Chloroplast (Reliability: 1)
NCED1_SOLLC
605
0
67317
Swiss-Prot
Chloroplast (Reliability: 2)
NCED2_ARATH
583
0
65067
Swiss-Prot
other Location (Reliability: 3)
NCED2_ORYSJ
576
0
63868
Swiss-Prot
Chloroplast (Reliability: 1)
NCED2_SOLLC
581
0
64834
Swiss-Prot
Chloroplast (Reliability: 4)
NCED3_ARATH
599
0
65857
Swiss-Prot
Chloroplast (Reliability: 3)
NCED3_ORYSJ
608
0
65651
Swiss-Prot
Mitochondrion (Reliability: 5)
NCED4_ORYSJ
582
0
62948
Swiss-Prot
Chloroplast (Reliability: 2)
NCED5_ARATH
589
0
65337
Swiss-Prot
Chloroplast (Reliability: 2)
NCED5_ORYSJ
613
0
65929
Swiss-Prot
Chloroplast (Reliability: 2)
NCED6_ARATH
577
0
63821
Swiss-Prot
Mitochondrion (Reliability: 5)
NCED9_ARATH
657
0
73015
Swiss-Prot
other Location (Reliability: 3)
NCED_ONCHC
610
0
67825
Swiss-Prot
Chloroplast (Reliability: 1)
A0A5B6YS12_DAVIN
640
0
69756
TrEMBL
Chloroplast (Reliability: 2)
A0A2I0B9Y5_9ASPA
968
0
106670
TrEMBL
Chloroplast (Reliability: 3)
A0A2P6Q9B1_ROSCH
612
0
67610
TrEMBL
Chloroplast (Reliability: 2)
A0A2I0A5Q3_9ASPA
594
0
65219
TrEMBL
Chloroplast (Reliability: 2)
A0A072V8S5_MEDTR
610
0
68302
TrEMBL
Chloroplast (Reliability: 1)
H6V2Y9_LACSA
578
0
64089
TrEMBL
Chloroplast (Reliability: 5)
A0A2I0AMZ3_9ASPA
551
0
60188
TrEMBL
Chloroplast (Reliability: 2)
A0A2G9FVR9_9LAMI
493
0
55611
TrEMBL
Chloroplast (Reliability: 5)
A0A2R4SC68_CAMSI
485
0
53893
TrEMBL
other Location (Reliability: 2)
H9U373_9ROSA
501
0
56395
TrEMBL
other Location (Reliability: 3)
A0A5B6Z6A7_DAVIN
288
0
32184
TrEMBL
other Location (Reliability: 5)
A0A5B6YYR6_DAVIN
182
0
19496
TrEMBL
Chloroplast (Reliability: 2)
A0A2I0B8J1_9ASPA
616
0
67005
TrEMBL
other Location (Reliability: 5)
A0A2R4SC72_CAMSI
615
0
66940
TrEMBL
Chloroplast (Reliability: 2)
K9N0S8_9BRAS
353
0
39757
TrEMBL
other Location (Reliability: 5)
A0A7G9MA00_AMAPA
319
0
36079
TrEMBL
other Location (Reliability: 2)
A0A2G9GNP4_9LAMI
601
0
67140
TrEMBL
Chloroplast (Reliability: 4)
A0A2G9HED4_9LAMI
462
0
51470
TrEMBL
Chloroplast (Reliability: 3)
H6V2Z1_9ASTR
578
0
64073
TrEMBL
Chloroplast (Reliability: 5)
W5RWJ6_9BRAS
259
0
29013
TrEMBL
other Location (Reliability: 2)
A0A5B6YQM4_DAVIN
535
0
58583
TrEMBL
Chloroplast (Reliability: 2)
A0A2H9ZR63_9ASPA
622
0
68304
TrEMBL
Chloroplast (Reliability: 2)
A0A2G9GN87_9LAMI
469
0
52704
TrEMBL
Chloroplast (Reliability: 4)
A0A2G9HG37_9LAMI
466
0
52158
TrEMBL
Chloroplast (Reliability: 4)
H6V2Z0_9ASTR
578
0
64089
TrEMBL
Chloroplast (Reliability: 5)
A0A2P6Q010_ROSCH
563
0
63046
TrEMBL
Secretory Pathway (Reliability: 4)
A0A2R4SC71_CAMSI
604
0
66710
TrEMBL
Chloroplast (Reliability: 1)
A0A0K0K5D7_BRANA
534
0
60410
TrEMBL
other Location (Reliability: 4)
A0A5B6YQA5_DAVIN
641
0
69924
TrEMBL
Chloroplast (Reliability: 3)
A0A0B2SP93_GLYSO
493
0
55512
TrEMBL
Chloroplast (Reliability: 3)
A0A2G9FWB1_9LAMI
571
0
63713
TrEMBL
Chloroplast (Reliability: 3)
A0A2R4SC59_CAMSI
606
0
66713
TrEMBL
Chloroplast (Reliability: 2)
A0A2R4SC70_CAMSI
606
0
67613
TrEMBL
Chloroplast (Reliability: 3)
A0A072V1Z3_MEDTR
586
0
66859
TrEMBL
Mitochondrion (Reliability: 5)
A0A2I0BE20_9ASPA
599
0
66325
TrEMBL
Chloroplast (Reliability: 3)
A0A385EI28_9ROSI
610
0
67287
TrEMBL
Chloroplast (Reliability: 1)
A0A5B6Z854_DAVIN
425
0
47428
TrEMBL
other Location (Reliability: 4)
A0A1I9RXM8_SENAL
380
0
41462
TrEMBL
other Location (Reliability: 2)
A0A396GMS1_MEDTR
608
0
67378
TrEMBL
Chloroplast (Reliability: 1)
A0A7C9EI78_OPUST
165
0
18541
TrEMBL
Mitochondrion (Reliability: 4)
A0A2P6Q004_ROSCH
330
0
37341
TrEMBL
Secretory Pathway (Reliability: 5)
H6V2Z2_9ASTR
578
0
64104
TrEMBL
Chloroplast (Reliability: 5)
S5CK78_9ROSA
606
0
66864
TrEMBL
Chloroplast (Reliability: 1)
A0A0F7L0S1_TRIRP
166
0
18350
TrEMBL
other Location (Reliability: 2)
A0A0B4SX54_CAMSI
68
0
7652
TrEMBL
other Location (Reliability: 2)
A0A2P6PHC3_ROSCH
303
0
34619
TrEMBL
Mitochondrion (Reliability: 4)
A0A0B4SWT3_CAMSI
181
0
19088
TrEMBL
Chloroplast (Reliability: 2)
A0A2P6QS16_ROSCH
613
0
67291
TrEMBL
Chloroplast (Reliability: 3)
H9CH57_DIOKA
485
0
54739
TrEMBL
other Location (Reliability: 4)
A0A2G9GUU5_9LAMI
595
0
66077
TrEMBL
Chloroplast (Reliability: 3)
A0A2H4W6Y9_HEVBR
251
0
28364
TrEMBL
other Location (Reliability: 2)
A0A2Z5CVL5_PHATN
609
0
67720
TrEMBL
Chloroplast (Reliability: 1)
A0A2P6Q5L4_ROSCH
688
0
75954
TrEMBL
other Location (Reliability: 5)
A0A075IEG5_CROSA
627
0
70127
TrEMBL
Mitochondrion (Reliability: 5)
A0A7C9ESV2_OPUST
594
0
66042
TrEMBL
Chloroplast (Reliability: 3)
H6V2Z3_9ASTR
578
0
64104
TrEMBL
Chloroplast (Reliability: 5)
A0A075W0Z7_9ROSA
607
0
66877
TrEMBL
-
A0A075W2E0_9ROSA
614
0
68374
TrEMBL
-
A0A0A7EBR7_LYCCN
608
0
67508
TrEMBL
-
A0A0M4UGS6_WHEAT
595
0
64828
TrEMBL
-
A0A1B3B582_WHEAT
591
0
64488
TrEMBL
-
A0A1B3B5A6_WHEAT
571
0
62462
TrEMBL
-
A0A220T631_CITRE
609
0
67656
TrEMBL
-
A0A6P4Q8G0_GOSAR
597
0
65479
TrEMBL
-
A9LM96_CISCR
87
0
9662
TrEMBL
-
A9XSC1_PRUPE
246
0
27900
TrEMBL
-
A9XSC2_9ROSI
247
0
27938
TrEMBL
-
B4XWB6_CUCME
246
0
27437
TrEMBL
-
C4MK80_MALHU
606
0
66903
TrEMBL
-
C6ZJY5_PRUPE
247
0
27566
TrEMBL
-
C6ZLC7_DIOKA
246
0
27709
TrEMBL
-
I6VH10_9POAL
608
0
65914
TrEMBL
other Location (Reliability: 1)
K9L7L3_ACAEB
545
0
61542
TrEMBL
other Location (Reliability: 1)
M1GDS4_WHEAT
615
0
66702
TrEMBL
-
M9SZN2_LILLO
603
0
66756
TrEMBL
-
Q1XHJ1_CITLI
569
0
63300
TrEMBL
-
Q1XHJ2_CITSI
569
0
63092
TrEMBL
-
Q1XHJ3_CITUN
569
0
63191
TrEMBL
-
Q1XHJ4_CITLI
587
0
65316
TrEMBL
-
Q1XHJ5_CITSI
587
0
65466
TrEMBL
-
Q1XHJ6_CITUN
587
0
65498
TrEMBL
-
Q2PHF9_LACSA
578
0
64061
TrEMBL
-
Q2PHG0_LACSA
583
0
64521
TrEMBL
-
Q2PHG1_LACSA
607
0
66737
TrEMBL
-
Q2PHG2_LACSA
585
0
65480
TrEMBL
-
Q460X8_9FABA
602
0
66857
TrEMBL
-
Q52QS5_ONCHC
242
0
27052
TrEMBL
-
Q52QS7_ONCHC
244
0
27333
TrEMBL
-
Q70KY0_ARAHY
601
0
66851
TrEMBL
-
Q8YXV3_NOSS1
Nostoc sp. (strain PCC 7120 / SAG 25.82 / UTEX 2576)
475
0
53744
TrEMBL
-
Q9AXZ3_PERAE
569
0
63078
TrEMBL
-
Q9FS24_VIGUN
612
0
67715
TrEMBL
-
Q9M3Z9_SOLTU
604
0
67288
TrEMBL
-
W8EAQ2_9ROSI
606
0
67005
TrEMBL
-
html completed




 results (
results ( results (
results ( top
top





