2.4.2.19: nicotinate-nucleotide diphosphorylase (carboxylating)
This is an abbreviated version!
For detailed information about nicotinate-nucleotide diphosphorylase (carboxylating), go to the full flat file.
Word Map on EC 2.4.2.19 
Reaction
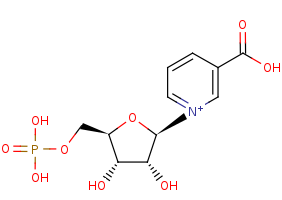 beta-nicotinate D-ribonucleotide +
beta-nicotinate D-ribonucleotide +
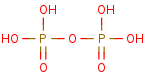 diphosphate +
diphosphate +
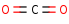 CO2=
CO2=
 pyridine-2,3-dicarboxylate +
pyridine-2,3-dicarboxylate +
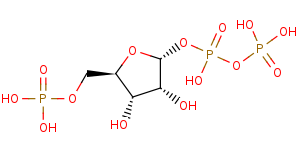 5-phospho-alpha-D-ribose 1-diphosphate
5-phospho-alpha-D-ribose 1-diphosphate
Synonyms
BNA6, general stress protein 70 , GSP70 , Hp-QAPRTase, hQPRTase, M6_Spy1061, NAD pyrophosphorylase, NadC, nicotinate mononucleotide pyrophosphorylase (carboxylating) (EC 2.4.2.19), nicotinate-nucleotide pyrophosphorylase (carboxylating), nicotinate-nucleotide:pyrophosphate phospho-alpha-D-ribosyltransferase (decarboxylating), pyrophosphorylase, nicotinate mononucleotide (carboxylating), QAPRTase, QPRT, QPRTase, QPRTase , QPT, QPT1, QPT2, quinolate phosphoribosyltransferase, quinolinate phosphoribosyl transferase, quinolinate phosphoribosyltansferase, quinolinate phosphoribosyltransferase, quinolinate phosphoribosyltransferase (decarboxylating), quinolinate phosphoribosyltransferase 1, quinolinate phosphoribosyltransferase 2, quinolinate phosphoribosyltransferase [decarboxylating] , quinolinic acid phosphoribosyl transferase, quinolinic acid phosphoribosyltransferase, quinolinic phosphoribosyltransferase, sp.NadC, spNadC, TM1645, type II quinolic acid phosphoribosyltransferase, type II quinolinic acid phosphoribosyltransferase
ECTree
Molecular Weight
Molecular Weight on EC 2.4.2.19 - nicotinate-nucleotide diphosphorylase (carboxylating)
Please wait a moment until all data is loaded. This message will disappear when all data is loaded.
Please wait a moment until the data is sorted. This message will disappear when the data is sorted.
167000
-
sucrose density gradient centrifugation
172000
-
sedimetation velocity method
178000
-
calculation from sedimentation and ultracentrifugation data
191000
6 * 191000, dynamic light scattering analysis
202000
-
equilibrium sedimentation
212300
dynamic light scattering
27500
-
8 * 27500, SDS-PAGE
30048
x * 30048, crystal structure, residues 1-273
31700
calculated from amino acid sequence
32000
-
5 * 32000, SDS-PAGE
33000
M1KCW7
6 * 33000, X-ray crystallography
33500
-
x * 33500, SDS-PAGE
34200
-
x * 34200, equilibrium sedimentation in guanidine HCl
36000
2 * 36000, SDS-PAGE
160000

-
gel filtration
160000
-
sucrose density gradient centrifugation, gel filtration
210000

-
gel filtration
34000

-
x * 34000, SDS-PAGE
34000
-
5 * 34000, SDS-PAGE
35000

-
2 * 35000, SDS-PAGE
35000
-
2 * 35000, SDS-PAGE
72000

-
gel filtration
72000
-
sucrose density gradient centrifugation




 results (
results ( results (
results ( top
top






