2.1.1.274: salicylate 1-O-methyltransferase
This is an abbreviated version!
For detailed information about salicylate 1-O-methyltransferase, go to the full flat file.
Word Map on EC 2.1.1.274 
Reaction
 S-adenosyl-L-methionine +
S-adenosyl-L-methionine +
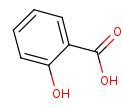 salicylate=
salicylate=
 S-adenosyl-L-homocysteine +
S-adenosyl-L-homocysteine +
 methyl salicylate
methyl salicylate
Synonyms
AlBSMT1, AtBSMT1, benzoate-salicylate methyltransferase, benzoic acid/SA methyltransferase, benzoic acid/salicylic acid carboxyl methyltransferase, benzoic acid/salicylic acid carboxyl methyltransferase 1, benzoic acid/salicylic acid methyltransferase, benzoic acid/salicylic acidcarboxyl methyltransferase, BSMT, BSMT1, class II SABATH protein, CsSAMT, LcSAMT, More, PaSABATH2, PaSAMT, PbBSMT methyltransferase, PtSABATH4, S-adenosyl-l-methionine: salicylic acid carboxyl methyl-transferase, S-adenosyl-L-methionine: salicylic acid carboxyl methyltransferase, S-adenosyl-L-methionine:salicylic acid carboxyl methyltransferase, SA carboxyl methyltransferase, SA methyl transferase, SA methyltransferase, SA MTase, SABATH methyltransferase 4, SABATH4, salicylic acid carboxyl methyltransferase, salicylic acid carboxyl methyltransferase-like, salicylic acid methyl transferase, salicylic acid methyltransferase, salicylicacid carboxyl methyl transferase, SAMT, SAMT1
ECTree
Sequence
Sequence on EC 2.1.1.274 - salicylate 1-O-methyltransferase
Please wait a moment until all data is loaded. This message will disappear when all data is loaded.
Please wait a moment until the data is sorted. This message will disappear when the data is sorted.
BSMT1_ARATH
379
0
43365
Swiss-Prot
other Location (Reliability: 2)
BSMT1_PETHY
357
0
40662
Swiss-Prot
other Location (Reliability: 2)
BSMT2_PETHY
357
0
40676
Swiss-Prot
other Location (Reliability: 2)
AAMT3_MAIZE
379
0
43220
Swiss-Prot
other Location (Reliability: 2)
SAMT_CLABR
359
0
40289
Swiss-Prot
other Location (Reliability: 3)
A0A2G9HE19_9LAMI
198
0
22683
TrEMBL
other Location (Reliability: 2)
A0A072UAJ0_MEDTR
357
0
40181
TrEMBL
other Location (Reliability: 2)
G7JWX2_MEDTR
421
0
48420
TrEMBL
other Location (Reliability: 4)
A0A088QFL1_ARTAN
374
0
41745
TrEMBL
other Location (Reliability: 3)
A0A072V8I3_MEDTR
369
0
42034
TrEMBL
other Location (Reliability: 4)
G7L6V5_MEDTR
357
0
40672
TrEMBL
other Location (Reliability: 2)
A0A396HL21_MEDTR
354
0
39849
TrEMBL
other Location (Reliability: 4)
A0A2P6PPD8_ROSCH
366
0
41201
TrEMBL
other Location (Reliability: 3)
A0A251S897_HELAN
371
0
41758
TrEMBL
other Location (Reliability: 2)
A0A088Q097_ARTAN
370
0
41843
TrEMBL
other Location (Reliability: 2)
A0A2G9HEC2_9LAMI
365
0
41672
TrEMBL
other Location (Reliability: 3)
A0A2G9HGR2_9LAMI
370
0
41409
TrEMBL
other Location (Reliability: 2)
A0A396HH47_MEDTR
129
0
14509
TrEMBL
other Location (Reliability: 5)
A0A2G9HE47_9LAMI
366
0
41309
TrEMBL
other Location (Reliability: 2)
B7FIK3_MEDTR
381
0
43579
TrEMBL
other Location (Reliability: 2)
G7L6V4_MEDTR
360
0
40870
TrEMBL
other Location (Reliability: 2)
A0A2P6PPF8_ROSCH
301
0
34246
TrEMBL
Chloroplast (Reliability: 4)
A0A396GLT2_MEDTR
180
0
20863
TrEMBL
Mitochondrion (Reliability: 2)
A0A167V6N5_CAMSI
367
0
41297
TrEMBL
other Location (Reliability: 2)
A0A2P6PPH0_ROSCH
366
0
41079
TrEMBL
other Location (Reliability: 2)
A0A396GNG7_MEDTR
362
0
40928
TrEMBL
other Location (Reliability: 3)
A0A2G9HGH3_9LAMI
365
0
41268
TrEMBL
other Location (Reliability: 2)
A0A396JH06_MEDTR
191
0
21599
TrEMBL
other Location (Reliability: 2)
A0A075W3C7_9LILI
360
0
41048
TrEMBL
-
A0A0A1E7P5_9LAMI
364
0
40673
TrEMBL
-
A0A514TTE2_LYCCN
357
0
39930
TrEMBL
-
A9PF83_POPTR
364
0
40693
TrEMBL
-
D2Y3T9_SOLLC
361
0
41235
TrEMBL
-
Q6XMI1_ARALL
149
0
17095
TrEMBL
-
Q8H6N2_ANTMA
383
0
43690
TrEMBL
-
R4I7S9_PLABS
377
0
42507
TrEMBL
-
html completed



 results (
results ( results (
results ( top
top









