2.1.1.157: sarcosine/dimethylglycine N-methyltransferase
This is an abbreviated version!
For detailed information about sarcosine/dimethylglycine N-methyltransferase, go to the full flat file.
Reaction
2 S-adenosyl-L-methionine
+
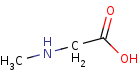 sarcosine=
2 S-adenosyl-L-homocysteine
+
sarcosine=
2 S-adenosyl-L-homocysteine
+
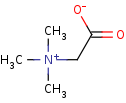 betaine
betaine
Synonyms
dimethylglycine methyltransferase, DMT, DMT2, glycine sarcosine dimethylglycine methyltransferase, glycine sarcosine methyltransferase, GSDMT, GsSDMT, MXAN 3190, sarcosine dimethylglycine methyltransferas, sarcosine dimethylglycine methyltransferase, sarcosine dimethylglycine N-methyltransferase, Sdm, Sdm homologue protein, SdmA, SDMT
ECTree
Sequence
Sequence on EC 2.1.1.157 - sarcosine/dimethylglycine N-methyltransferase
Please wait a moment until all data is loaded. This message will disappear when all data is loaded.
Please wait a moment until the data is sorted. This message will disappear when the data is sorted.
GSDMT_ACTHA
565
0
64719
Swiss-Prot
-
SDMT_APHHA
277
0
31613
Swiss-Prot
-
SDMT_HALHR
279
0
32228
Swiss-Prot
-
A0A428C1B3_STROR
254
0
29338
TrEMBL
-
A0A136MTF0_9BACT
236
0
27263
TrEMBL
-
A0A5B9W0H4_9BACT
269
0
29912
TrEMBL
-
A0A5B9WE72_9BACT
262
0
28902
TrEMBL
-
A0A7Y9DXS1_9PSEU
564
0
63434
TrEMBL
-
A0A3S4SG26_9ACTO
284
0
31741
TrEMBL
-
A0A518AGS2_9BACT
277
0
30745
TrEMBL
-
A0A0P1IBS8_9RHOB
273
0
30081
TrEMBL
-
A0A3S4VAT7_MYCCI
208
0
22957
TrEMBL
-
A0A428DY76_STRMT
254
0
29358
TrEMBL
-
A0A1B8YJW0_9GAMM
274
0
30745
TrEMBL
-
A0A4P6Q2U1_9ACTN
Streptomonospora litoralis
216
0
23427
TrEMBL
-
A0A5P9GN48_9RHOB
359
0
40522
TrEMBL
-
A0A0P1EAF1_9RHOB
259
0
28567
TrEMBL
-
A0A1U7MAZ5_9FIRM
255
0
29005
TrEMBL
-
A0A2X1SG85_MYCXE
243
0
26570
TrEMBL
-
A0A2D5IVZ4_9RHOB
274
0
29471
TrEMBL
-
K9YGK5_HALP7
Halothece sp. (strain PCC 7418)
277
0
31613
TrEMBL
-
A0A3S4GXK2_MORMO
260
0
28736
TrEMBL
-
A0A0Y3LEP5_STREE
254
0
29333
TrEMBL
-
A0A0T9QBF3_9GAMM
265
0
30150
TrEMBL
-
A0A445NA81_STRNE
291
0
32373
TrEMBL
-
A0A518HPL3_9BACT
278
0
30976
TrEMBL
-
A0A1Q2HP91_9BACT
277
0
31237
TrEMBL
-
A0A4V0ER07_STROR
254
0
29430
TrEMBL
-
A0A428G0Y6_STROR
254
0
29307
TrEMBL
-
A0A6J4R3J4_9ACTN
277
0
32154
TrEMBL
-
A0A822IZP1_9EURY
274
0
30638
TrEMBL
-
A0A2X2P9L0_STRGR
279
0
30621
TrEMBL
-
A0A6N3I3C4_9CLOT
253
0
28534
TrEMBL
-
H2CAA9_9LEPT
276
0
31431
TrEMBL
-
A0A1C5UKA8_9FIRM
uncultured Blautia sp
256
0
29049
TrEMBL
-
A0A7W9YK21_9ACTN
543
0
62178
TrEMBL
-
A0A2X3XL78_STRSA
254
0
29509
TrEMBL
-
A0A162FIW6_9EURY
300
0
35441
TrEMBL
-
A0A1V5RVA7_9CHLR
257
0
28597
TrEMBL
-
A0A428C481_STROR
254
0
29165
TrEMBL
-
A0A5P9NS79_9BACT
148
0
16671
TrEMBL
-
A0A2C9CBD5_KUEST
290
0
33148
TrEMBL
-
A0A2T7YJ74_BACTU
253
0
29109
TrEMBL
-
A0A1W7AET3_9STAP
243
0
28132
TrEMBL
-
A0A1V5BC74_9FIRM
245
0
27701
TrEMBL
-
A0A2G9FYU0_9LAMI
145
0
16406
TrEMBL
other Location (Reliability: 3)
A0A1V6K4I8_9BACT
266
0
29662
TrEMBL
-
A0A259V1I9_9FIRM
255
0
28883
TrEMBL
-
A0A3R9NEQ9_STRSA
254
0
29407
TrEMBL
-
A0A3G4VQV6_9ACTN
282
0
31087
TrEMBL
-
A0A0P1E443_9RHOB
259
0
28594
TrEMBL
-
A0A655EJR9_MYCTX
50
0
5729
TrEMBL
-
A0A1Y5TED8_9RHOB
Roseovarius litorisediminis
275
0
29394
TrEMBL
-
A0A2X2WHI4_CITKO
256
0
28468
TrEMBL
-
A0A5S9QSN4_9GAMM
296
0
32728
TrEMBL
-
A0A1V5URL9_9BACT
274
0
31359
TrEMBL
-
A0A380XZI6_BACCE
240
0
28407
TrEMBL
-
A0A173TLY0_9FIRM
256
0
29049
TrEMBL
-
A0A1V2RQU7_9ACTN
292
0
31593
TrEMBL
-
A0A852YZ57_9ACTN
565
0
64851
TrEMBL
-
A0A378TLZ5_9MYCO
206
0
22766
TrEMBL
-
A0A7W6W927_9PROT
582
0
65461
TrEMBL
-
A0A7W7PHP1_STRNE
285
0
32018
TrEMBL
-
A0A0W0RK34_LEGBO
334
0
37506
TrEMBL
-
A0A853AP89_9PSEU
567
0
64520
TrEMBL
-
A0A1D8FYW2_9ACTN
277
0
30555
TrEMBL
-
A0A1C5NN43_9CLOT
uncultured Clostridium sp
226
0
25791
TrEMBL
-
A0A1V4I9G4_9CLOT
217
0
25723
TrEMBL
-
A0A3R9M3L1_STROR
254
0
29431
TrEMBL
-
A0A0F7KNX7_9SPHN
274
0
30626
TrEMBL
-
A0A1Y5RNH8_9RHOB
259
0
28275
TrEMBL
-
A0A0G3EG49_9BACT
279
0
31739
TrEMBL
-
A0A1V6JIH3_9BACT
207
0
22907
TrEMBL
-
A0A6A7SEY3_9ZZZZ
268
0
30707
TrEMBL
other Location (Reliability: 1)
A0A839DSA9_9PSEU
Halosaccharopolyspora lacisalsi
569
0
64916
TrEMBL
-
A0A1Y2NYY1_STRFR
277
0
30555
TrEMBL
-
A0A5P9G8L8_9RHOB
195
0
21261
TrEMBL
-
A0A2T7YM11_BACTU
209
0
24169
TrEMBL
-
A0A3P1S1S1_STRSA
254
0
29411
TrEMBL
-
A0A6B3L3B2_9BACI
207
0
23849
TrEMBL
-
A0A2N8NTQ4_STREU
283
0
31047
TrEMBL
-
A0A2X4NBC6_9BACL
254
0
29362
TrEMBL
-
A0A1Y2MUQ1_PSEAH
209
0
21896
TrEMBL
-
A0A7Y9XX82_9SPHN
274
0
29956
TrEMBL
-
A0A380YFU3_BACCE
209
0
24268
TrEMBL
-
A0A7W0HM04_9DELT
278
0
31764
TrEMBL
-
A0A5C6BRU4_9PLAN
275
0
30553
TrEMBL
-
A0A840NKI6_9PSEU
569
0
64932
TrEMBL
-
A0A428DGY6_STRMT
254
0
29221
TrEMBL
-
A0A3R9HVC1_STRSA
254
0
29382
TrEMBL
-
A0A3N6ERX9_9ACTN
282
0
31073
TrEMBL
-
A1WXM9_HALHL
Halorhodospira halophila (strain DSM 244 / SL1)
278
0
32010
TrEMBL
-
A0A6N2V0L6_9BACT
244
0
27706
TrEMBL
-
A0A448KJW2_HAEDC
251
0
28041
TrEMBL
-
A0A840E980_9BACT
274
0
31972
TrEMBL
-
A0A231I2A4_BACTU
249
0
29056
TrEMBL
-
A0A3N9FN43_9BURK
320
0
36290
TrEMBL
-
A0A0D6JI85_9HYPH
278
0
31143
TrEMBL
-
A0A2X1TJH7_MORMO
260
0
28672
TrEMBL
-
A0A380EW13_9STAP
205
0
23123
TrEMBL
-
A0A4V0BUN9_9STRE
254
0
29320
TrEMBL
-
A0A1Y2MMQ6_PSEAH
276
0
29860
TrEMBL
-
A0A1Y2MMQ6_PSEAH
276
0
29860
TrEMBL
-
A0A840QD78_9PSEU
567
0
64605
TrEMBL
-
A0A3R9LUP4_STRCR
254
0
29455
TrEMBL
-
A0A428HMN9_STRCR
254
0
29214
TrEMBL
-
A0A3R9ILA5_STRMT
254
0
29303
TrEMBL
-
A0A444IWI0_9DELT
277
0
32727
TrEMBL
-
A0A428A0Z5_STRSA
254
0
29293
TrEMBL
-
A0A378UQT8_9MYCO
223
1
24663
TrEMBL
-
A0A5B1CEU1_9BACT
279
0
31337
TrEMBL
-
A0A5B9QPR9_9BACT
298
0
32383
TrEMBL
-
A0A5C6CLG4_9BACT
282
0
31537
TrEMBL
-
A0A839XKG5_9PSEU
566
0
64024
TrEMBL
-
A0A6N2WKB7_9CLOT
uncultured Clostridium sp
207
0
23266
TrEMBL
-
A0A1V5RGC9_9CHLR
260
0
28855
TrEMBL
-
A0A5K1I0T4_9GAMM
Halomonas lysinitropha
286
0
30772
TrEMBL
-
A0A498PQN1_9MYCO
426
0
46749
TrEMBL
-
A0A498PA86_9MYCO
278
0
30400
TrEMBL
-
A0A5B9MFU0_9BACT
282
0
31446
TrEMBL
-
A0A1V2RV53_9ACTN
208
0
22402
TrEMBL
-
A0A3E2YKW5_9ACTN
224
0
24095
TrEMBL
-
A0A655JHJ0_MYCTX
103
0
11781
TrEMBL
-
A0A3R9SSE4_STRCR
254
0
29306
TrEMBL
-
A0A427ZW40_STRSA
254
0
29351
TrEMBL
-
A0A3N6G802_9ACTN
308
0
33322
TrEMBL
-
A0A4R8SZX3_9MYCO
249
0
27348
TrEMBL
-
A0A427Z128_STROR
254
0
29350
TrEMBL
-
A0A6V6ZNC0_9RHOB
259
0
28406
TrEMBL
-
A0A1E3XE42_9BACT
268
0
30958
TrEMBL
-
A0A3N6F0T4_9ACTN
270
0
29543
TrEMBL
-
A0A5J4E4U0_9BACT
284
0
31223
TrEMBL
-
A0A839S4N2_9PSEU
566
0
63901
TrEMBL
-
A0A852TT51_9ACTN
552
0
63188
TrEMBL
-
A0A3R9LGB0_STROR
254
0
29421
TrEMBL
-
A0A139REN5_STRMT
254
0
29360
TrEMBL
-
A0A045IXC8_MYCTX
116
0
13059
TrEMBL
-
A0A380NCU0_STRGR
308
0
33590
TrEMBL
-
A0A387HPH5_9ACTN
283
0
31529
TrEMBL
-
A0A174FVW0_9FIRM
238
0
27404
TrEMBL
-
A0A5C6E524_9BACT
278
0
31499
TrEMBL
-
A0A5C6FP93_9PLAN
282
0
31520
TrEMBL
-
A0A5B7UR00_9ACTN
298
0
32996
TrEMBL
-
A0A7W7N117_9ACTN
568
0
64856
TrEMBL
-
A0A5B8RAQ5_9ZZZZ
279
0
31605
TrEMBL
other Location (Reliability: 1)
A0A7W6S159_9PROT
559
0
63550
TrEMBL
-
A0A259UCU3_9FIRM
249
0
27999
TrEMBL
-
A0A841EBH0_9ACTN
520
0
59439
TrEMBL
-
A0A399HBZ1_9ACTN
295
0
32185
TrEMBL
-
A0A5P2XLR5_STRST
286
0
31030
TrEMBL
-
A0A078LEK5_CITKO
256
0
28438
TrEMBL
-
A0A0H5BEB9_BLAVI
251
0
27179
TrEMBL
-
F4Y2P8_9CYAN
Moorena producens 3L
278
0
31999
TrEMBL
-
A0A3R9MW71_STROR
254
0
29350
TrEMBL
-
A0A080M2B3_9PROT
262
0
29076
TrEMBL
-
A0A2S8BNE9_9MYCO
210
0
22934
TrEMBL
-
A0A852W6Q9_9PSEU
278
0
30153
TrEMBL
-
A0A852W5J2_9PSEU
283
0
32729
TrEMBL
-
A0A853BNE0_9ACTN
521
0
59570
TrEMBL
-
S3JE43_MICAE
279
0
32185
TrEMBL
-
A0A387HQJ0_9ACTN
279
0
30545
TrEMBL
-
A0A109CNC4_9BACT
Candidatus Magnetominusculus xianensis
345
0
39448
TrEMBL
-
A0A1Y2NIT2_STRPT
251
0
27674
TrEMBL
-
A0A656HC91_THINJ
Thiothrix nivea (strain ATCC 35100 / DSM 5205 / JP2)
278
0
30608
TrEMBL
-
A0A395LD62_9HYPH
269
0
29952
TrEMBL
-
A0A3S3SLA5_9DELT
283
0
32382
TrEMBL
-
A0A045J2Z4_MYCTX
101
0
11710
TrEMBL
-
A0A498QJI9_9MYCO
282
0
30835
TrEMBL
-
A0A2R8B3H2_9RHOB
289
0
31075
TrEMBL
-
A0A7W8QR53_9ACTN
552
0
62892
TrEMBL
-
A0A3R9KFK1_STRCR
254
0
29349
TrEMBL
-
A0A3N6HNC9_9ACTN
279
0
30614
TrEMBL
-
A0A132C378_9RHOB
214
0
24084
TrEMBL
-
A0A174FH57_9FIRM
397
0
46227
TrEMBL
-
D5EA51_METMS
Methanohalophilus mahii (strain ATCC 35705 / DSM 5219 / SLP)
278
0
31504
TrEMBL
-
A0A7X5ZQ20_9PSEU
569
0
64454
TrEMBL
-
A0A3N6DWU5_9ACTN
270
0
29646
TrEMBL
-
A0A2S6TVT0_9PROT
279
0
31003
TrEMBL
-
A0A1W6LPW9_9BACT
277
0
31249
TrEMBL
-
A0A100JYE1_STRSC
335
0
36522
TrEMBL
-
A0A852ZGN2_9ACTN
558
0
62921
TrEMBL
-
A0A2K8REC8_9ACTN
208
0
22006
TrEMBL
-
A0A1Y2MI42_STRPT
273
0
30091
TrEMBL
-
A0A1C5XEI1_9CLOT
uncultured Clostridium sp
201
0
22783
TrEMBL
-
A0A428CC28_STROR
254
0
29256
TrEMBL
-
A0A109ZX66_9DELT
287
0
31797
TrEMBL
-
A0A2P9FG19_9ACTN
311
0
33296
TrEMBL
-
A0A448PBW9_9ACTO
265
0
29291
TrEMBL
-
A0A2T7YN98_BACTU
239
0
27491
TrEMBL
-
A0A433E191_9ACTN
282
0
30835
TrEMBL
-
A0A5C5Y7G6_9PLAN
282
0
31251
TrEMBL
-
A0A8F8UEK1_9ACTN
406
0
43425
TrEMBL
-
A0A1D2ICG1_9ACTN
276
0
30540
TrEMBL
-
A0A1V5UI97_9BACT
277
0
31321
TrEMBL
-
A0A448QQD6_ACTEU
218
0
24628
TrEMBL
-
A0A1Y6AS32_9BACI
209
0
24180
TrEMBL
-
A0A1C5W7E8_9CLOT
uncultured Clostridium sp
226
0
25830
TrEMBL
-
A0A399GL47_9ACTN
222
0
24130
TrEMBL
-
A0A081R4H5_STROR
254
0
29275
TrEMBL
-
A0A0H5P2D1_NOCFR
208
0
22586
TrEMBL
-
A0A2S6UE41_9PROT
277
0
30647
TrEMBL
-
A0A0N8NTS3_9CLOT
232
0
26461
TrEMBL
-
A0A428APC2_STRCR
254
0
29438
TrEMBL
-
A0A3R9MPL6_STRCR
254
0
29282
TrEMBL
-
A0A428CD34_STRMT
254
0
29250
TrEMBL
-
A0A3N6H593_9ACTN
308
0
33649
TrEMBL
-
A0A3R9I029_STRSA
254
0
29388
TrEMBL
-
A0A142YCR6_9PLAN
270
0
29780
TrEMBL
-
A0A839S3T8_9PSEU
283
0
31807
TrEMBL
-
A0A3R9IFK5_STROR
254
0
29417
TrEMBL
-
A0A2X0R822_9LACT
244
0
27946
TrEMBL
-
A0A2H5ZNQ7_9BACT
243
0
27738
TrEMBL
-
A0A7W7R987_KITKI
279
0
30672
TrEMBL
-
A0A5B8REV5_9ZZZZ
268
0
30707
TrEMBL
other Location (Reliability: 1)
A0A6N2SF87_9CLOT
uncultured Clostridium sp
226
0
25834
TrEMBL
-
A0A1V4WTF9_9DELT
228
0
25196
TrEMBL
-
A0A136WHB4_9FIRM
246
0
28219
TrEMBL
-
A0A5K1I0J0_9GAMM
Halomonas lysinitropha
218
0
24155
TrEMBL
-
A0A8B3IZS9_STROR
254
0
29264
TrEMBL
-
A0A2P9EZ45_9ACTN
252
0
27847
TrEMBL
-
A0A1V4IJN4_9CLOT
259
0
30369
TrEMBL
-
A0A1U9NLV9_9BACT
279
0
31467
TrEMBL
-
A0A238KYT2_9RHOB
252
0
27833
TrEMBL
-
A0A2S6X4S0_9ACTN
208
0
22327
TrEMBL
-
A0A2S6QLJ0_9PROT
275
0
31031
TrEMBL
-
A0A0P1I549_9RHOB
259
0
28537
TrEMBL
-
A0A5B7V4X8_9ACTN
284
0
30665
TrEMBL
-
A0A428GI93_STRCR
254
0
29382
TrEMBL
-
A0A1K2FU97_9ACTN
297
0
32981
TrEMBL
-
A0A8D9P4P5_9FIRM
397
0
46181
TrEMBL
-
A0A2S6TKX2_9PROT
283
0
30411
TrEMBL
-
A0A5S9Q5Q2_9GAMM
296
0
32820
TrEMBL
-
A0A822J361_9EURY
269
0
30055
TrEMBL
-
A0A377G0Z9_9BACL
231
0
25405
TrEMBL
-
A0A2S6TBI5_9PROT
285
0
32099
TrEMBL
-
A0A2R8B3M1_9RHOB
258
0
28337
TrEMBL
-
A0A1X6ZKY4_9RHOB
267
0
29943
TrEMBL
-
A0A378WR51_9NOCA
269
0
29250
TrEMBL
-
A0A2S6SPX5_9PROT
261
0
29283
TrEMBL
-
A0A7W7W0R3_9ACTN
551
0
63022
TrEMBL
-
A0A1V5Y592_9BACT
261
1
29974
TrEMBL
-
A0A1B8YHR0_9GAMM
314
0
35875
TrEMBL
-
A0A7H4LUV7_9ENTR
228
0
25408
TrEMBL
-
A0A2H5X4Q3_9BACT
278
0
30665
TrEMBL
-
A0A2X1TE44_MYCXE
274
0
29455
TrEMBL
-
A0A109UKR8_9GAMM
218
0
23999
TrEMBL
-
A0A822J1U8_9EURY
292
0
32730
TrEMBL
-
A0A5B9P7L5_9BACT
260
0
29075
TrEMBL
-
A0A2H6FVF7_9BACT
166
0
18611
TrEMBL
-
A0A377I2A8_HAEPH
251
0
28093
TrEMBL
-
A0A498PTN2_9MYCO
296
0
32192
TrEMBL
-
A0A6A7SGE9_9ZZZZ
279
0
31605
TrEMBL
other Location (Reliability: 1)
A0A2Z4UJM2_9HYPH
272
0
29145
TrEMBL
-
A0A3R9GQR4_STRSA
254
0
29348
TrEMBL
-
A0A2K8PNX5_STRLA
282
0
31096
TrEMBL
-
F6KV62_9EURY
278
0
31620
TrEMBL
-
html completed




 results (
results ( results (
results ( top
top





